View libraries and data sets used in these notes
# load packages library (tidyverse) library (tidymodels) library (knitr) library (patchwork)library (GGally) # for pairwise plot matrix library (corrplot) # for correlation matrix # load data <- read_csv ("https://sta221-fa25.github.io/data/rail_trail.csv" )# set default theme in ggplot2 :: theme_set (ggplot2:: theme_bw ())
Topics
Multicollinearity
Definition
How it impacts the model
How to detect it
What to do about it
Data: Trail users
The Pioneer Valley Planning Commission (PVPC) collected data at the beginning a trail in Florence, MA for ninety days from April 5, 2005 to November 15, 2005 to
Data collectors set up a laser sensor, with breaks in the laser beam recording when a rail-trail user passed the data collection station.
Source: Pioneer Valley Planning Commission via the mosaicData package.
Variables
Outcome :
volume estimated number of trail users that day (number of breaks recorded)
Predictors
hightemp daily high temperature (in degrees Fahrenheit)
avgtemp average of daily low and daily high temperature (in degrees Fahrenheit)
season one of “Fall”, “Spring”, or “Summer”
precip measure of precipitation (in inches)
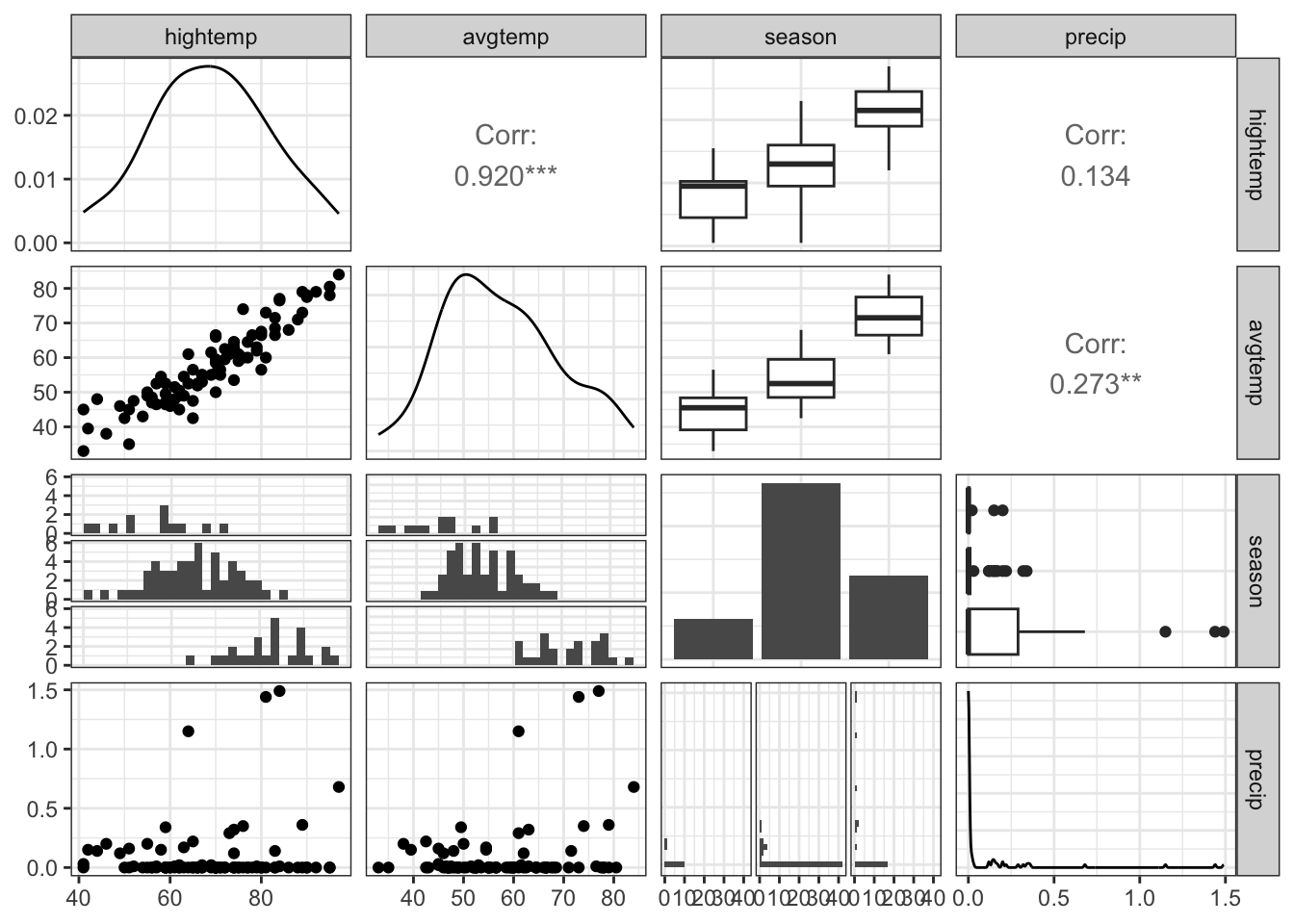
EDA: Relationship between predictors
We can create a pairwise plot matrix using the ggpairs function from the GGally R package
|> select (hightemp, avgtemp, season, precip) |> ggpairs ()
EDA: Relationship between predictors
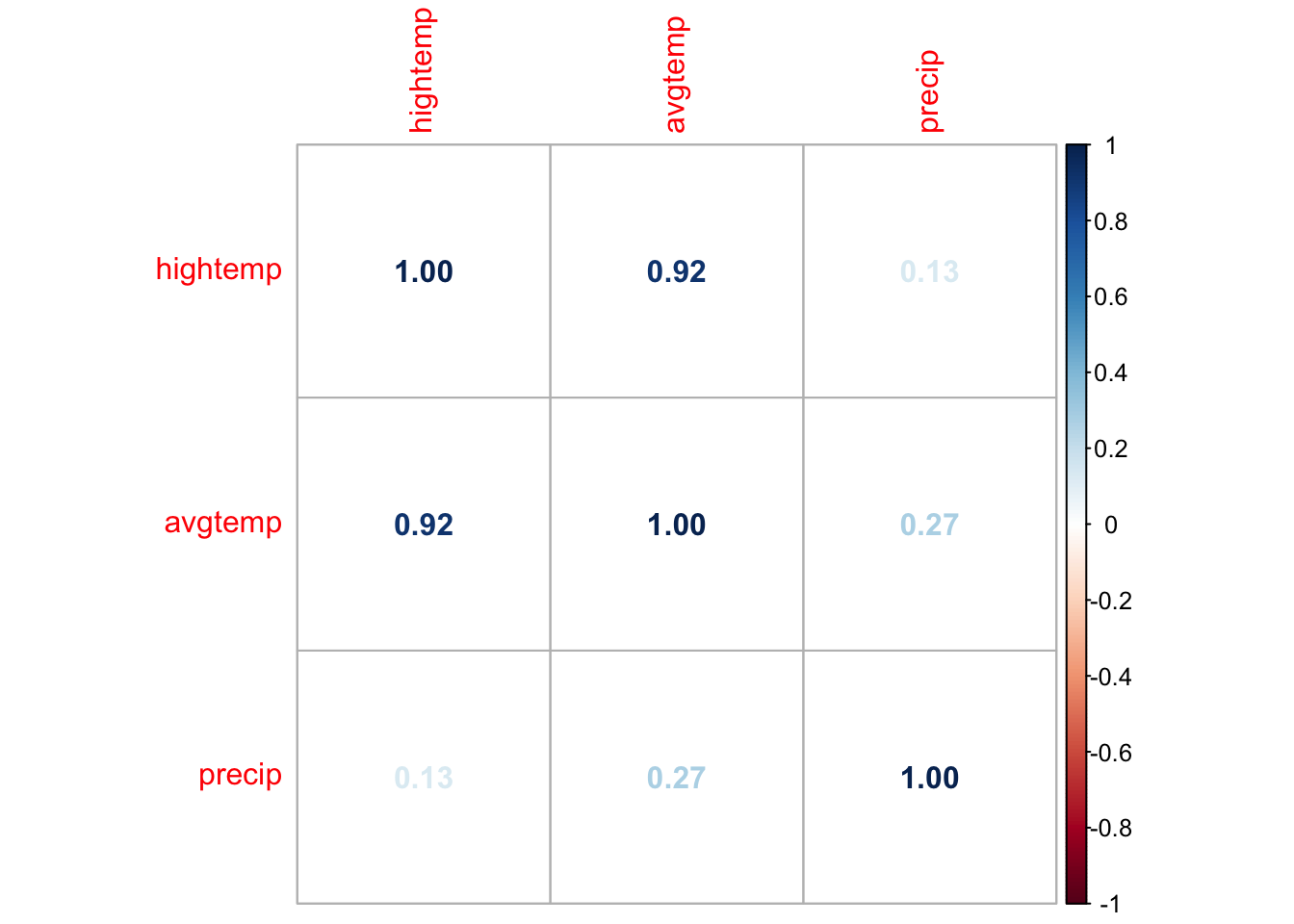
EDA: Correlation matrix
We can. use corrplot() in the corrplot R package to make a matrix of pairwise correlations between quantitative predictors
<- rail_trail |> select (hightemp, avgtemp, precip) |> cor ()corrplot (correlations, method = "number" )
EDA: Correlation matrix
What might be a potential concern with a model that uses high temperature, average temperature, season, and precipitation to predict volume?
Multicollinearity
Multicollinearity
Ideally the predictors are orthogonal, meaning they are completely independent of one another
In practice, there is typically some dependence between predictors but it is often not a major issue in the model
If there is linear dependence among (a subset of) the predictors, we cannot find estimate \(\hat{\boldsymbol{\beta}}\)
If there are near-linear dependencies, we can find \(\hat{\boldsymbol{\beta}}\) but there may be other issues with the model
Multicollinearity : near-linear dependence among predictors
Sources of multicollinearity
Data collection method - only sample from a subspace of the region of predictors
Constraints in the population - e.g., predictors family income and size of house
Choice of model - e.g., adding high order terms to the model
Overdefined model - have more predictors than observations
Detecting multicollinearity
How could you detect multicollinearity?
Variance inflation factor
The variance inflation factor (VIF) measures how much the linear dependencies impact the variance of the predictors
\[
VIF_{j} = \frac{1}{1 - R^2_j}
\]
where \(R^2_j\) is the proportion of variation in \(x_j\) that is explained by a linear combination of all the other predictors
When the response and predictors are scaled in a particular way, \(C_{jj} = VIF_{j}\) . Click here to see how.
Relationship with variance
Recall that \(Var(\hat{\beta}) = \sigma^2 (X^TX)^{-1}\) . If we call the matrix \((X^TX)^{-1}\) “C” then the variance of \(\hat{\beta}_j\) is \(\sigma^2 C_{jj}\) . Specifically,
\[
C_{jj} = VIF \times \frac{1}{\sum (X_{ij} - \bar{X_j})^2}
\]
Detecting multicollinearity
Common practice uses threshold \(VIF > 10\) as indication of concerning multicollinearity (some say VIF > 5 is worth investigation)
Variables with similar values of VIF are typically the ones correlated with each other
Use the vif() function in the rms R package to calculate VIF
library (rms)<- lm (volume ~ hightemp + avgtemp + precip, data = rail_trail)vif (trail_fit)
hightemp avgtemp precip
7.161882 7.597154 1.193431
Load the data using the code below:
<- read_csv ("https://sta221-fa25.github.io/data/rail_trail.csv" )
and compute VIF manually (without using the function vif()).
How multicollinearity impacts model
Large variance for the model coefficients that are collinear
Different combinations of coefficient estimates produce equally good model fits
Unreliable statistical inference results
May conclude coefficients are not statistically significant when there is, in fact, a relationship between the predictors and response
Interpretation of coefficient is no longer “holding all other variables constant”, since this would be impossible for correlated predictors
Dealing with multicollinearity
Collect more data (often not feasible given practical constraints)
Redefine the correlated predictors to keep the information from predictors but eliminate collinearity
e.g., if \(x_1, x_2, x_3\) are correlated, use a new variable \((x_1 + x_2) / x_3\) in the model
For categorical predictors, avoid using levels with very few observations as the baseline
Remove one of the correlated variables
Be careful about substantially reducing predictive power of the model
Recap
End of class notes
library (tidyverse)library (tidymodels)<- read_csv ("https://sta221-fa25.github.io/data/rail_trail.csv" )<- lm (hightemp ~ avgtemp + precip, data = rail_trail)1 / (1 - glance (model1)$ r.squared)set.seed (221 )= initial_split (rail_trail, prop = 3 / 4 )= training (rt_split)= testing (rt_split)<- lm (volume ~ hightemp + avgtemp + precip, data = rt_train)<- lm (volume ~ avgtemp + precip, data = rt_train)<- rt_test$ volume - predict (model_full, newdata = rt_test)<- rt_test$ volume - predict (model_minus_temp, newdata = rt_test)## Compare root mean squared error (RMSE) sqrt (sum (residual_full^ 2 ))sqrt (sum (residual_minus_temp^ 2 ))## Examine coefficients for each model